Welcome to the Ramp Atlas
Quickly identify ramp sequences in your human and SARS-CoV-2 genes of interest.
Ramp sequences can have a large affect on gene expression. We present extensive analyses of ramp sequences in human and SARS-CoV-2 genes, across tissues and cell types. The Ramp Atlas is a website that enables users to quickly calculate ramp sequences in small fasta files. Users can also download data in convenient CSV files about the prevalence of ramp sequences in genes calculated from stratified tissue and cell types. Enjoy!
Pages
Try ExtRamp Online
Run the powerful ExtRamp program from a simple user-friendly interface, allowing you to quickly identify ramp sequences in genes without having to touch a line of code.
Search the Data
Whether a gene has a ramp sequence depends on local tRNA abundancies in the specific tissue or cell type. Query genes, tissues, and cell types here to learn more about where ramp sequences are found.
SARS-CoV-2
Ramp sequences are found in several genes in SARS-CoV-2, potentially affcting tissue-specific infection. Click to learn more about SARS-CoV-2 ramp sequences!
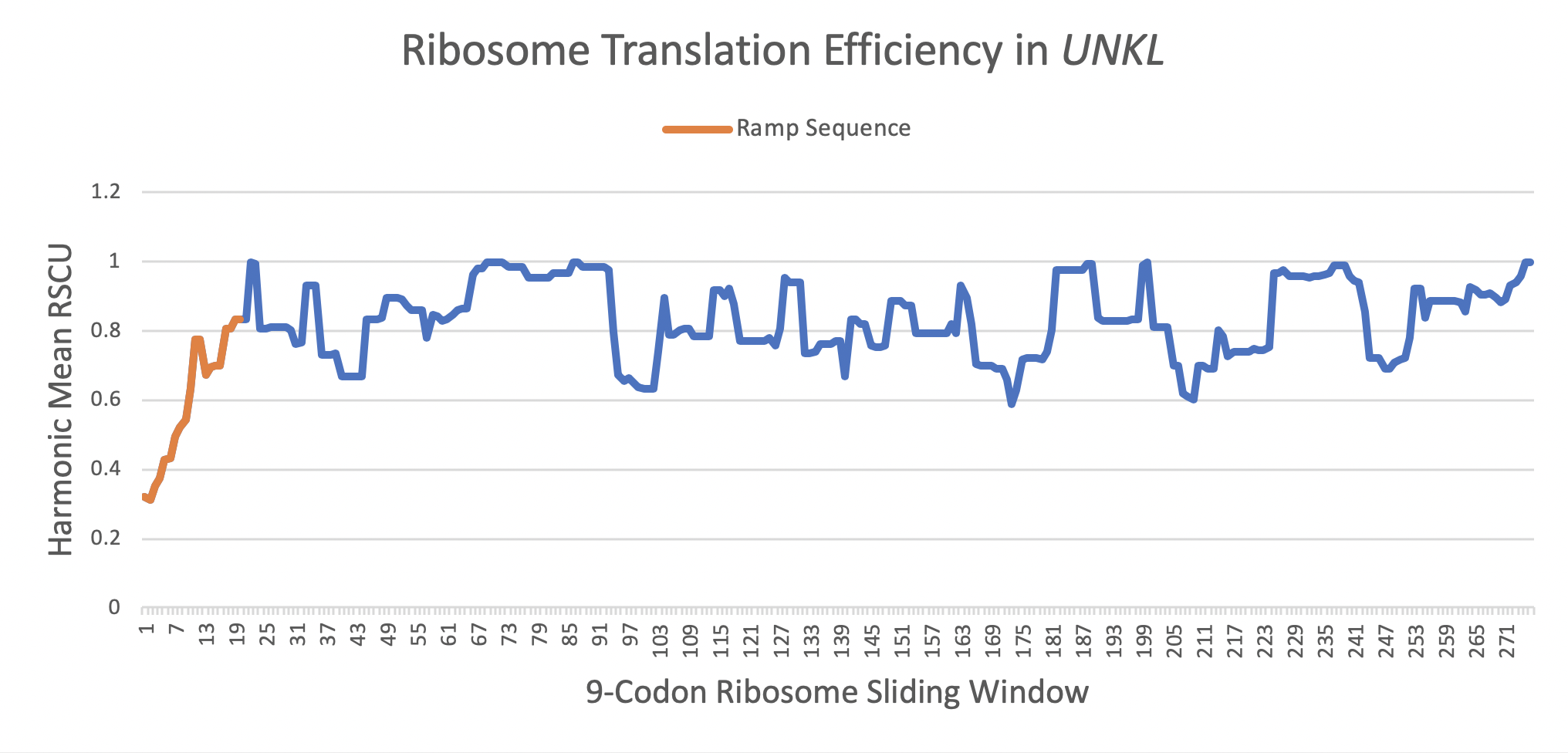
What is a ramp sequence?
A ramp sequence is a segment of rare, slowly translated codons at the beginning of a gene.
This altered initiation of translation increases gene expression by evenly spacing ribosomes and reducing mRNA secondary structure. The graph below shows the translational efficiency (measured as a Relative Synonymous Codon Usage, or RSCU, value) of codons throughout the gene. The orange segment is the ramp sequence which has a mean translational efficiency of 0.4531, while the entire gene has a mean efficiency of 0.7734. A translational efficiency of 1.0 would be the most rapidly translated.
Additional Links
Read the Paper
The original ExtRamp paper describes ramp sequences in detail as well as exactly how ExtRamp identifies them. This is a great place to go for a more in-depth understanding.
Visit the GitHub
For full access to the features of ExtRamp, download the python program from our GitHub page. ExtRamp.py can be run from the command line on UNIX machines.